Privacy-Preserving and Energy-Efficient MONAI Medical Image Annotation with NVIDIA Holoscan Edge Compute
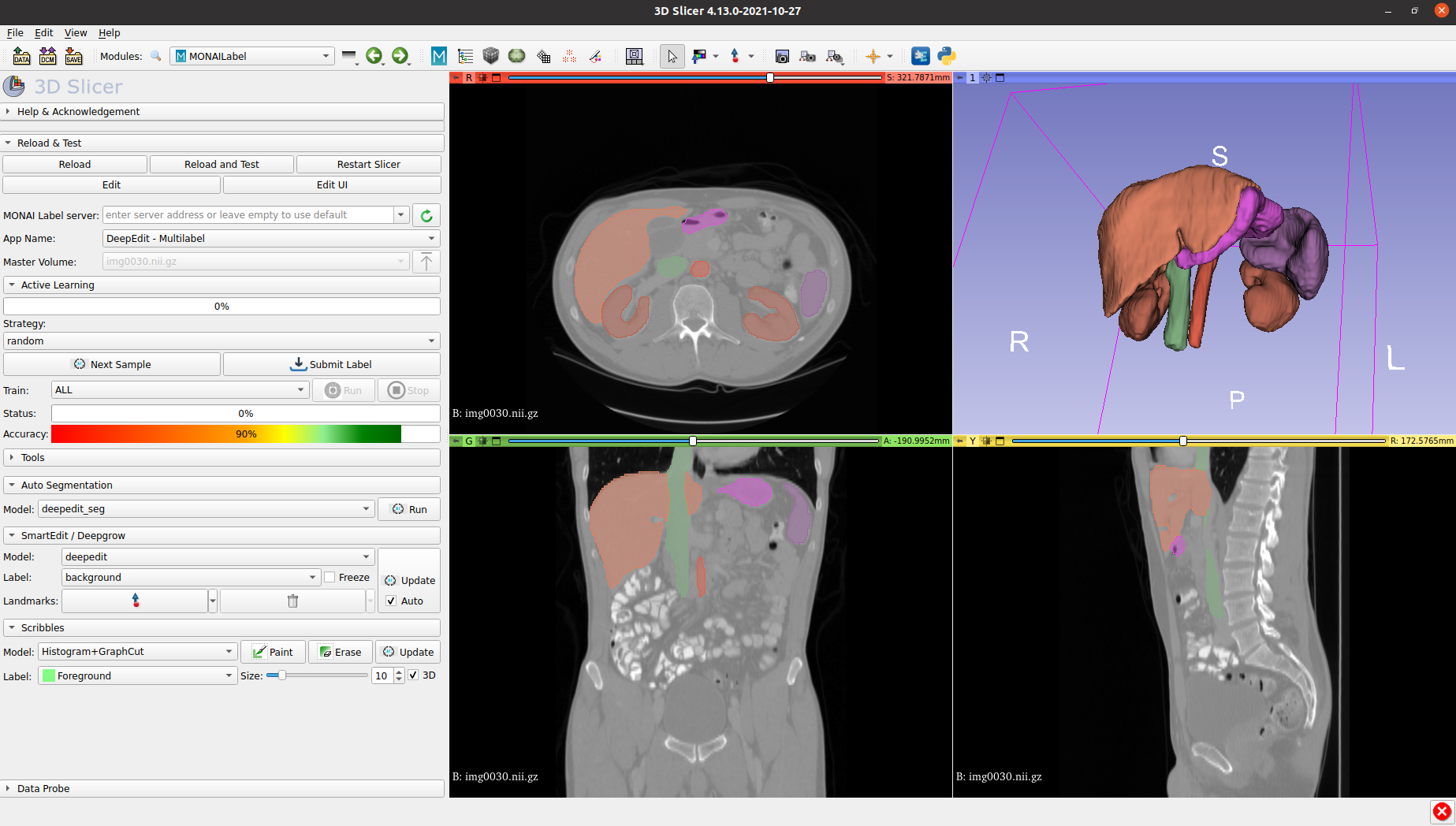
The collection and preparation of healthcare data is a prerequisite to any development of AI applications. Optimizing annotation pipelines is critical because the evolution of AI model architectures and the imperative to make them generalize well for production involves more and more data being collected. As such, the annotation stage often becomes the bottleneck. For annotation workflows to keep up with the pace of data collection, it is important to deploy end-to-end optimized pipelines. In addition, in the medical domain, it is important to ensure privacy, security, and energy-efficiency of the hardware and process for it to be adopted in healthcare settings. This is where edge and embedded compute platforms such as NVIDIA IGX Developer Kits can shine.
Among other annotation problems, whole-body CT segmentation task is notoriously challenging both for annotators and reviewers, specifically:
- How to handle centralized secure data storage and synchronization
- How to enable fast interaction with the data
- How to accelerate annotation speed and improve accuracy
In collaboration with the NVIDIA healthcare team, we propose a workflow based on MONAI Label, using 3D Slicer as a client to tackle these challenges. We profiled the usage of the compute resource with a focus on energy consumption and computational efficiency.
Setup MONAI Label workflow on NVIDIA Clara AGX DevKit
MONAI is an open source platform designed to address the challenges of integrating deep learning into healthcare solutions. Inside the MONAI ecosystem lives MONAI Label, which focuses on the task of scalable and efficient medical image labeling. For more information on MONAI workflows, we invite the reader to check this post.
Here, we focused on spleen segmentation on the well-known medical decathlon dataset. The test dataset was annotated with a client-server paradigm, where the MONAI Label server is running on the NVIDIA Clara AGX Developer Kit, and the user interacts with the 3D Slicer client.
Platform used
NVIDIA Holoscan, optimized on the Clara AGX Developer Kit, is a computing architecture for next-generation AI medical devices. It is optimized for the Holoscan framework and system software, a multimodal real-time AI sensor streaming platform allowing devices to evolve from fixed function to software-defined.
The embedded system-on-chip (SoC) is a Jetson AGX Xavier module embedding an 8-core Carmel ARM v8.2 CPU with an integrated Volta GPU (512 CUDA and 64 Tensor cores), and also an RTX 6000 discrete GPU. The ARM-based CPU, widely used in mobile devices, allows for energy-efficient embedded applications. More information on the Developer Kit can be found in the technical specs.

Software requirements
With HoloPack, it is quite easy to setup the Clara AGX Developer Kit. It embeds all APIs like CUDA and cudnn needed to develop AI applications. Here, we updated the firmware to 5.10.65-tegra on Ubuntu 20.04.4 LTS.
MONAI Label server and client setup
For the server, we used monailabel 0.8.3, monai 1.3.2 and pytorch 2.2 with upgraded cuda-12.2. On the client-side, we used 3D Slicer 5.6.2, with the MONAI Label extension.
Check the following documentation for setup instructions.
Methods
We annotated all the test dataset from MSD on the CLARA AGX Developer Kit with the RTX 6000 discrete GPU and compared it with a desktop including an 12th Gen Intel i9-12900K and a similar GPU (NVIDIA RTX A6000) on Ubuntu 22.04 LTS.
For the annotation workflow, we first made sure that all the data was cached by the Slicer client in ~/.cache/slicer.org/Slicer/SlicerIO. After running the radiology app server on the selected device, we simulated an annotator workflow by loading each CT volume with the random strategy, and sending the auto-segmented label to the data storage server.
While running the application, we profiled the auto-segmentation time along with the GPU and CPU consumption with the following custom commands:
- GPU NVIDIA (desktop & AGX)
sudo nvidia-smi -lms 100 nvidia-smi -lms 100 --query-gpu=timestamp,gpu_name,driver_version,utilization.gpu,memory.used,memory.total,utilization.memory,temperature.gpu,power.draw.instant,power.max_limit --format=csv - CPU Intel (desktop)
sudo turbostat -i 0.1 -s CoreTmp,Busy%,PkgWatt -S - CPU Tegra (AGX)
sudo tegrastats --interval 100
For an overview of the application running, please look at the following video:
Results
A summary table of the results is shown in Table 1.
| Metric \ Device | Desktop (RTX A6000) | Clara AGX (RTX 6000) |
| Prediction time (s) | 1.8 | 4.2 |
| CPU consumption mean/max (W) | 4.44 / 118.5 | 0.97 / 3.87 |
| CPU temperature mean/max (°C) | 28.68 / 60.0 | 38.28 / 40.5 |
| GPU consumption mean/max (W) | 58.5 / 283.7 | 54.76 / 266.96 |
| GPU temperature mean/max (°C) | 51.3 / 67.0 | 41.21 / 52.0 |
| GPU VRAM mean/max (GB) | 3.96 / 12.12 | 4.6 / 12.44 |
Although the prediction time is twice as long compared to that on a desktop workstation due to the higher-end RTX GPU on the workstation, the CPU consumption is really impressive thanks to the reduced instruction-set from the ARM CPU. When its AI compute capacity is sufficient on the iGPU, we could limit the use to the iGPU instead of the discrete RTX GPU (dGPU) to further reduce the consumption to below 30W. The temperatures are also lower which is overall more secure for regulatory considerations.
Conclusion
Throughout this post, we showed that using a Clara AGX Developer Kit in the context of medical AI can improve the overall workflow and provide a powerful all-in-one platform for federated annotations.
We expect more developments to be done on the platform, especially on the state-of-the-art NVIDIA Orin IGX platform in the near-future so stay tuned!
Don’t hesitate to contact us for any inquiries or questions.
Acknowledgments
We want to thank NVIDIA for providing us a Clara AGX Developer Kit.